About
Wearable data are becoming an essential source of health and disease data as they inform on various personal, behavioral, social, contextual, and environmental health-relevant factors. Wearables have been primarily used for activity tracking and gained popularity with fitness applications; however, more recently, these devices have been used in an increasing number of health applications, including health monitoring, clinical care, remote clinical trials, drug delivery, and disease characterization, to name a few.
We linked wearable data to clinical outcomes, and we have found that data from wearable measurements are as good or better than clinical measurements for predicting adverse health events in cancer medicine, diagnosing neuromotor disorders in infants, and for cardiac monitoring.
Our research on wearable data is conducted in partnership with a group of community stakeholders of medical teams that use wearable data in various clinical applications. The resulting research products include open-source software made available to the larger health community as part of the W4H Toolkit.
W4H Toolkit
The W4H Toolkit consists of an open source software built a modern analytical platform to work efficiently with wearable data. The W4H analytical platform supports data collection, storage, indexing, query, and analytics for many wearable data types, such as activity, heart rate, acceleration and gyroscope data, and demographic information. The platform is designed from the ground up to enable spatiotemporal data processing to leverage cases where location information is available with the the wearable sensor data. The platform also includes components to work with streaming data for real-time analytics.
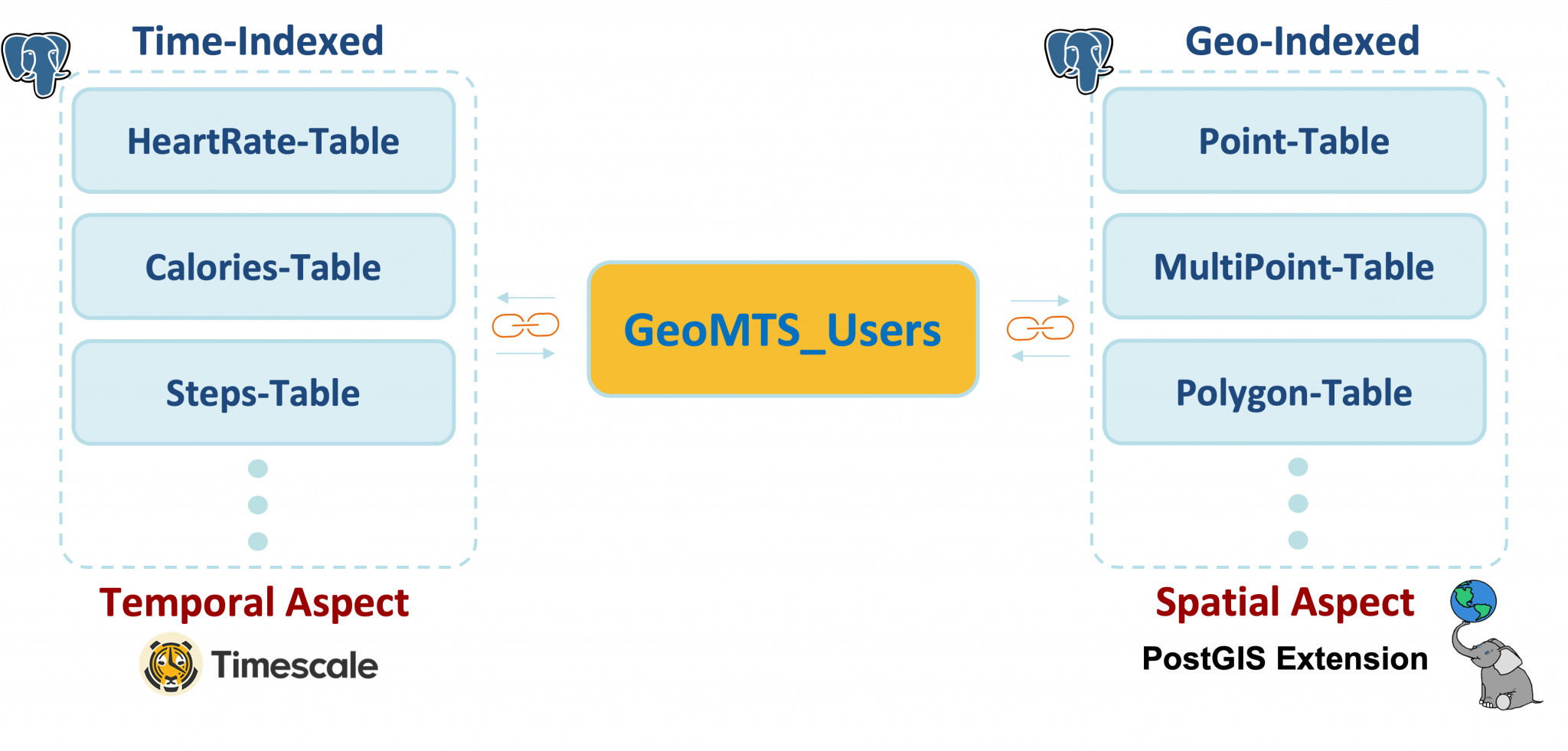
To enable spatiotemporal analytics, we conceptualize wearable data as Geo-referenced Multivariate Time Series (GeoMTS) data, i.e., time series with geotagged information. This conceptualization allows us to represent different types of wearable data in a single form to separate the wearable type from its data management and analysis.
We have implemented the W4H analytical platform on Spark, a popular open-source big data platform. To enable the research community to use the W4H platform with their data and sensors we have released the W4H Toolkit, a suite of extensible open-source software packages that include the W4H platform, utilities, and add-on components.
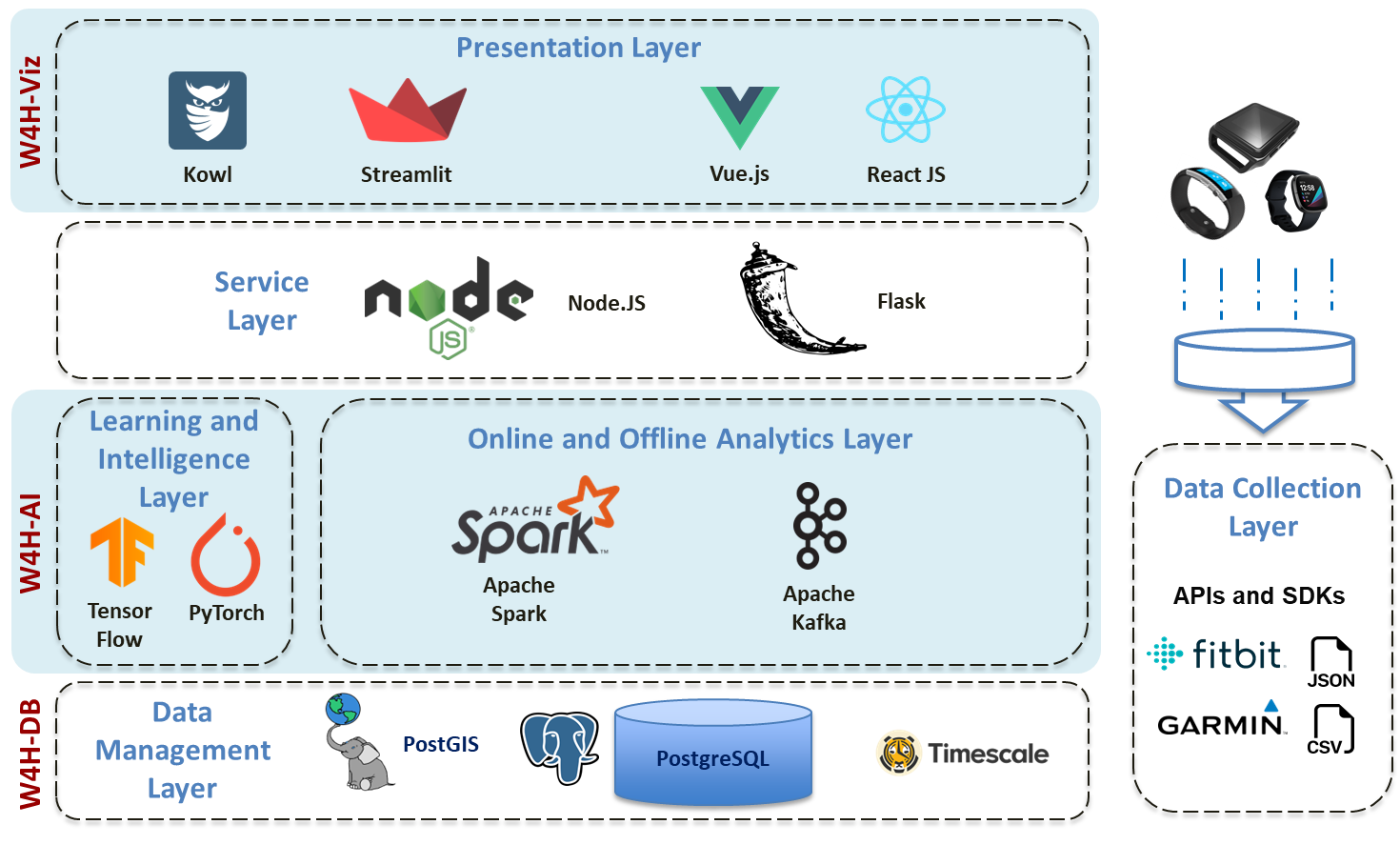
W4H Toolkit packages
StreamSim
StreamSim is a real-time data streaming simulator tool that allows researchers and developers to simulate real-time streaming behavior for tabular data. It provides a convenient way to test real-time applications when the actual real-time data source is unavailable or when simulating different scenarios for testing purposes. GitHub repository
pyGarminAPI
pyGarminAPI is a Python library that provides a convenient way to interact with the Garmin API through the Garmin Pull API. The library allows you to send signed requests, authenticate with your Garmin API credentials, and fetch data using Garmin Pull API for your Garmin devices. GitHub repository
Case Studies
These case studies showcase the application of the W4H Toolkit on various data and sensors.
Cancer Patient Monitoring
We analyze cancer patients under IRB protocol OS-16-2 at USC who were undergoing treatment with chemotherapy to study their fatigue level. Our results show that ECOG score did not predict the rate of adverse events, such as unexpected hospitalization and unplanned trips to the day hospital for hydration. By comparison, determination of the patient’s calorie expenditure during waking hours was highly predictive of serious adverse events over the course of the study.
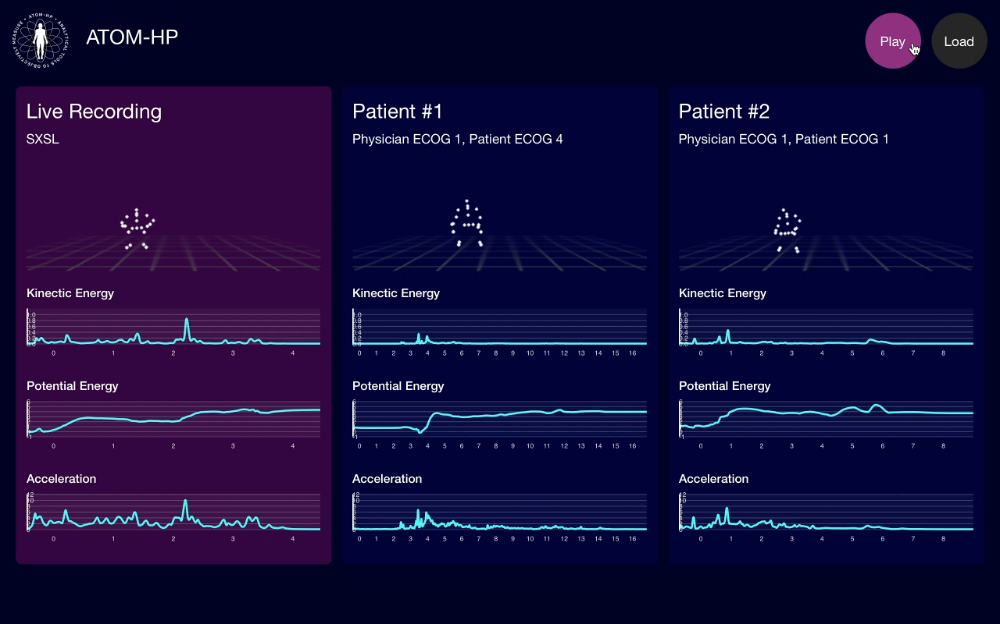
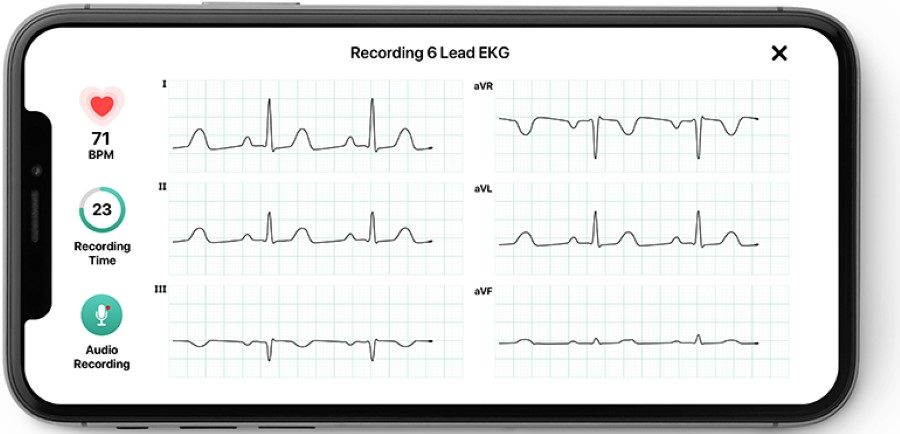
Cardiac Monitoring
Atrial fibrillation (AF) affects 1% of the general population and causes a third of all strokes. Smartphone ECG has the potential to overcome the shortcomings of traditional ECG recorders. The results have demonstrated the unique capabilities to expand the diagnostic scope, allowing patients to nimbly monitor for arrhythmia instantly on demand and outside of a traditional healthcare setting. Our further data analysis not only validates the ability of devices to accurately detect cardiac rhythms, but also demonstrates a consumer appetite for a device that enables continuous monitoring.
- Infant Neuromotor Monitoring: Early identification of impaired neuromotor control in infancy is necessary in order to provide early therapy intervention in accordance with known principles of experience-dependent neuroplasticity. Early intervention is often not initiated until an infant has demonstrated a consistent and severe delay in the acquisition of developmental milestones, in contrast with known principles of experience-dependent neuroplasticity. We have recently validated the use of wearable sensors for unobtrusive full-day, in-home movement monitoring of spontaneous infant leg movements.

References
- Luan Tran, Yanfang Li, Luciano Nocera, Cyrus Shahabi, and Li Xiong, MultiFusionNet: Atrial Fibrillation Detection With Deep Neural Networks, AMIA 2020 Information Summit , Houston, Texas, March 23-26, 2020.
- Tanachat Nilanon, Luciano P. Nocera, Alexander S. Martin, Anand Kolatkar, Marcella May, Zaki Hasnain, Naoto T. Ueno, Sriram Yennu, Angela Alexander, Aaron E. Mejia, Roger Wilson Boles, Ming Li, Jerry S. H. Lee, Sean E. Hanlon, Frankie A. Cozzens Philips, David I. Quinn, Paul K. Newton, Joan Broderick, Cyrus Shahabi, Peter Kuhn, and Jorge J. Nieva, Use of Wearable Activity Tracker in Patients With Cancer Undergoing Chemotherapy: Toward Evaluating Risk of Unplanned Health Care Encounters, JCO Clinical Cancer Informatics, Sep 24, 2020.
- Luan Tran, Manh Nguyen, and Cyrus Shahabi, Representation Learning for Early Sepsis Prediction, 2019 Computing in Cardiology (CinC), Singapore, 8-11 Sept. 2019.
- Mohammad Saeed Abrishami, Luciano Nocera, Melissa Mert, Ivan A. Trujillo-Priego, Sanjay Purushotham, Cyrus Shahabi, and Beth A. Smith, Identification of Developmental Delay in Infants using Wearable Sensors: Full-Day Leg Movement Statistical Feature Analysis, IEEE Journal of Translational Engineering in Health and Medicine, 25 January 2019.
People
Student(s)
 |  |
| Arash Hajisafi CS PhD Student, USC | Maria Despoina Siampou CS PhD Student, USC |
Collaborator(s)
 |  |  |
| Peter Kuhn Keck School of Medicine, USC | Jorge Nieva Keck School of Medicine, USC | Leslie Saxon Keck School of Medicine, USC |
 |  |  |
| Beth Smith Keck School of Medicine, USC | Luciano Nocera Viterbi School of Engineering, USC | Yao-Yi Chiang Department of CSE, UMN |
Principal Investigator(s)
 |
| Cyrus Shahabi Viterbi School of Engineering, USC |
Former Members
 |  | |
| Alireza Abdoli CS PostDoc, USC | Asmita Chotani CS Master Student, USC |



